Laura Zeugner
Wissenschaftlerin
MPI für Marine Mikrobiologie
Celsiusstr. 1
D-28359 Bremen
|
Raum: |
1339 |
|
Telefon: |

MY RESEARCH
I'm working on my PhD thesis as part of the FACS Group, supervised by PD Dr. Bernhard Fuchs.
Visualization of cell-specific traits in marine picoplankton populations
The aim of my thesis is to optimize the methodology of direct-geneFISH and develop the applicability from isolates towards environmental samples. Furthermore, different high-resolution microscopy methods, such as Airyscan, structured illumination microscopy and stimulated emission depletion microscopy are evaluated for their ability to visualize FISH methods.
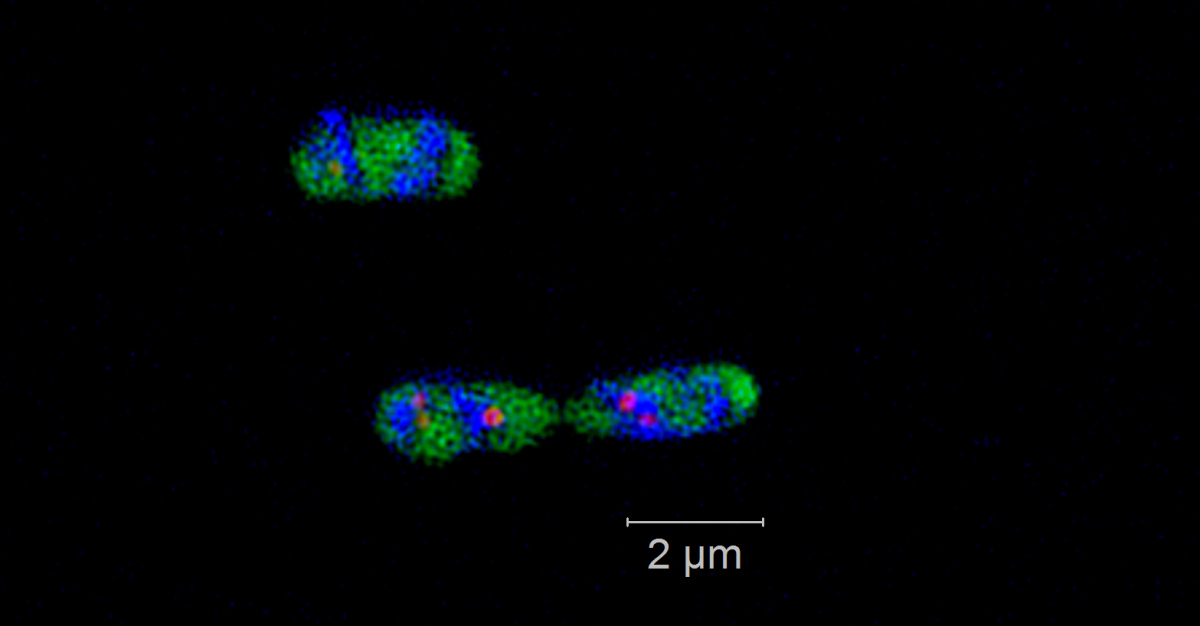
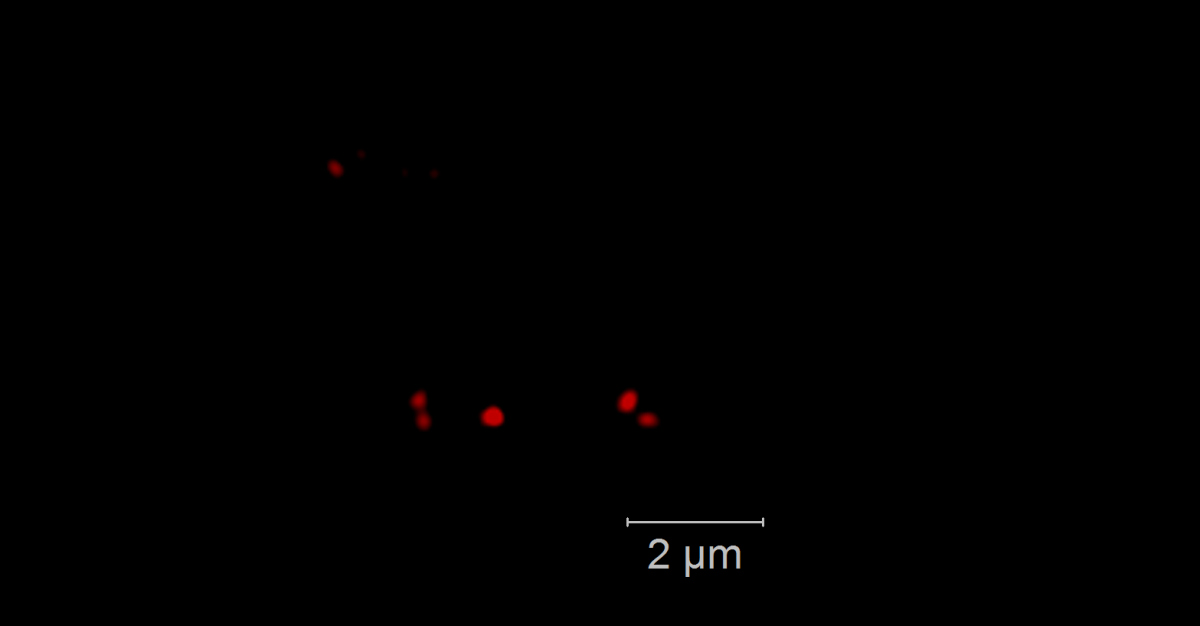
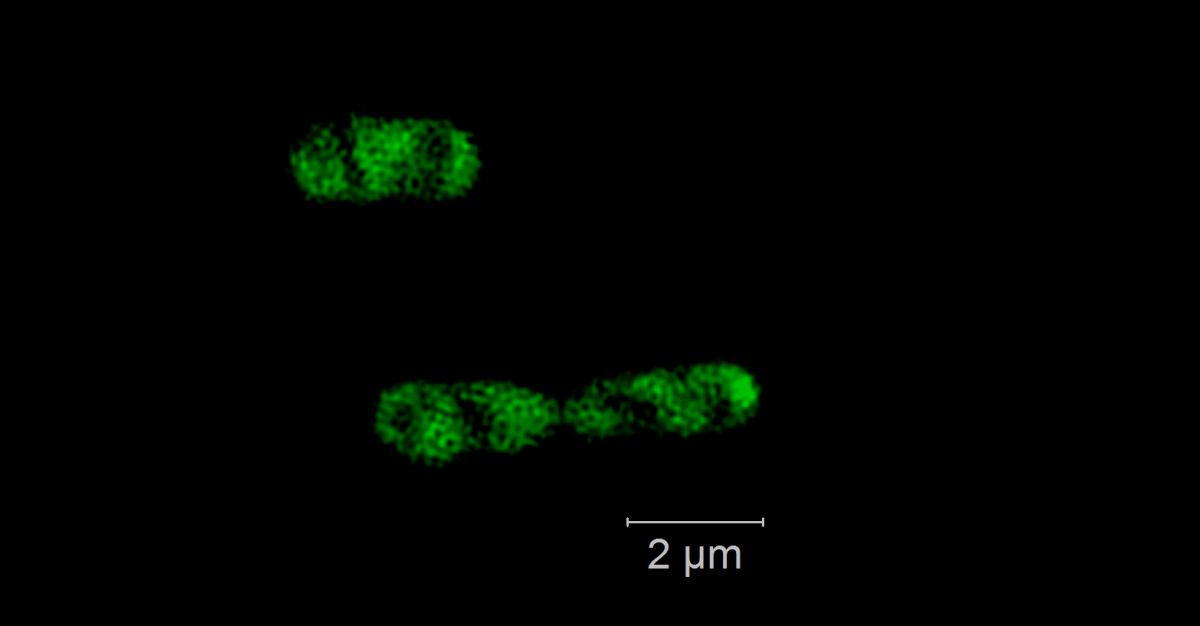
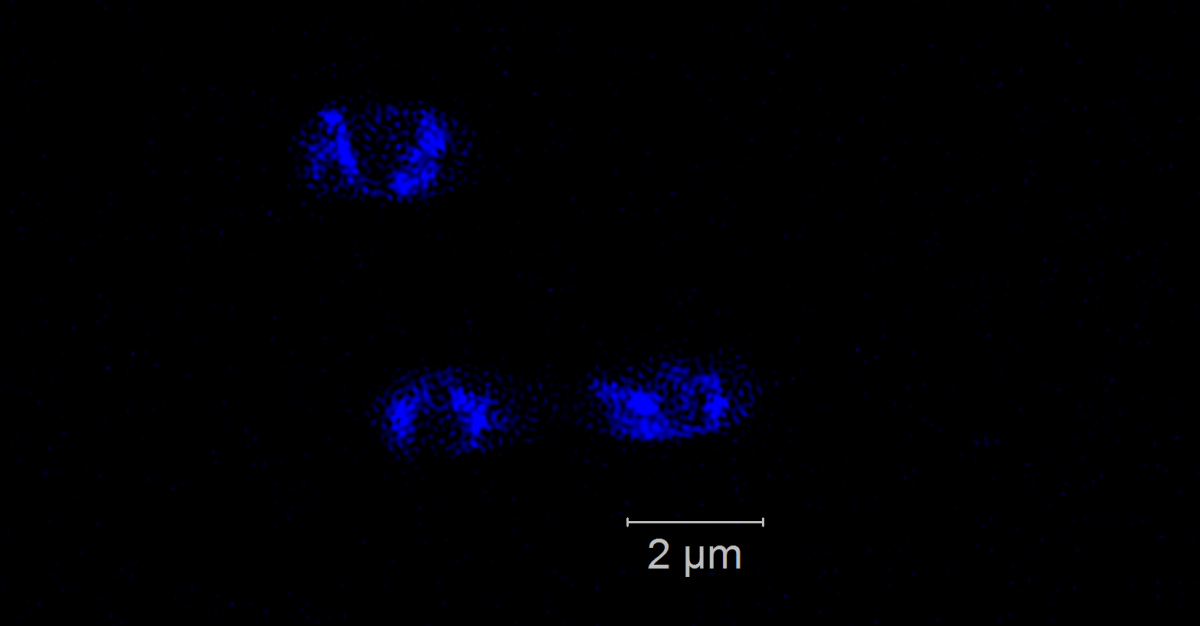
Phytoplankton blooms occur as seasonal events in many coastal areas. They grow within weeks to high biomasses and their decay is followed by an annually recurring succession of specialized heterotrophic bacteria. Genomic and proteomic analyses revealed that this succession is substrate-controlled and that certain bacterial strains have specific enzymatic repertoires for algal derived polysaccharide uptake and utilization [1]. Formosa strain B (Hel1_33_131) and Polaribacter (Hel1_33_49), two Flavobacteriia occurring during the algae spring bloom in the German Bight, have specialized polysaccharide utilization loci for the degradation of mannose-rich sulfated polysaccharides that are common compounds of diatom cell walls. Within those loci we want to detect and visualize specific glycoside hydrolase genes of family 92 that are coding for α-mannosidase. We use direct-geneFISH [2] to simultaneously detect the ribosomal RNA for phylogenetic affiliation and the gene of interest in the target organisms and different high-resolution microscopy methods to enable a subcellular localization of the gene signals in situ.
Our experiments show that it is possible to simultaneously visualize functional genes and their host’s rRNA on a single cell level not only in pure cultures but also in environmental samples containing populations of the flavobacterial species. By linking the presence of key genes involved in polysaccharide degradation with the cell identity on environmental samples, direct-geneFISH provides a quantitative insight into niche adaption, e.g. the ability to directly compete with other bacterioplankton clades during and after spring blooms. This method will prove especially valuable for investigating bacterioplankton populations based on their functional traits throughout the ocean.
[1] Teeling et al. (2012): Substrate-controlled succession of marine bacterioplankton populations induced by a phytoplankton bloom. Science
[2] Barrero-Canosa et al. (2016): Direct-geneFISH: a simplified protocol for the simultaneous detection and quantification of genes and rRNA in microorganisms. Environmental Microbiology
Direct geneFISH on E. coli cells