- Press Office
- Press releases 2021
- New form of symbiosis discovered
New form of symbiosis discovered
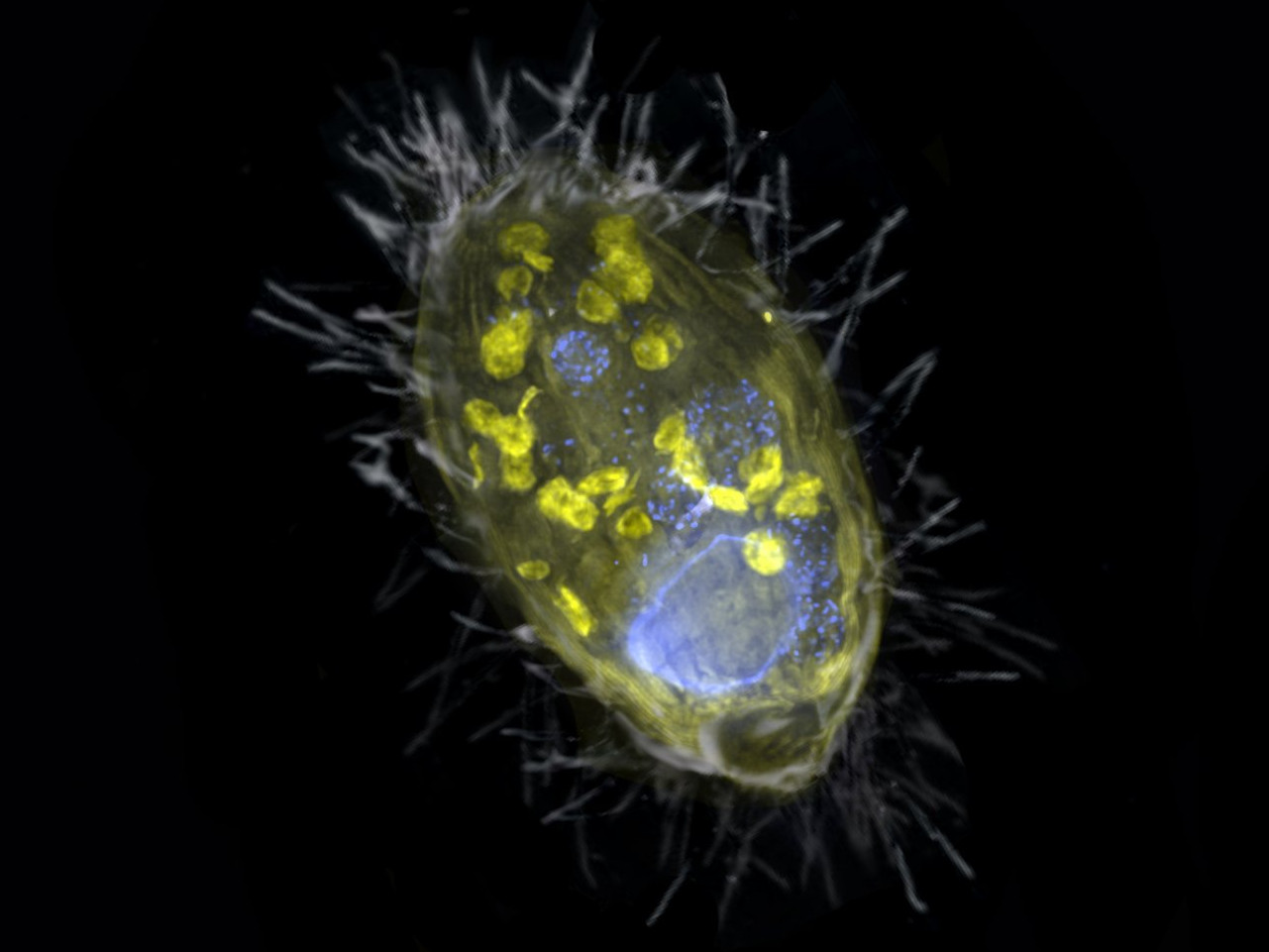
Researchers from Bremen, together with their colleagues from the Max Planck Genome Center in Cologne and the aquatic research institute Eawag from Switzerland, have discovered a unique bacterium that lives inside a unicellular eukaryote and provides it with energy. Unlike mitochondria, this so-called endosymbiont derives energy from the respiration of nitrate, not oxygen. “Such partnership is completely new,” says Jana Milucka, the senior author on the Nature paper. “A symbiosis that is based on respiration and transfer of energy is to this date unprecedented”.
In general, among eukaryotes, symbioses are rather common. Eukaryotic hosts often co-exist with other organisms, such as bacteria. Some of the bacteria live inside the host cells or tissue, and perform certain services, such defense or nutrition. In return, the host provides shelter and suitable living conditions for the symbiont. An endosymbiosis can even go that far that the bacterium loses its ability to survive on its own outside its host.
This was also the case with the symbiosis discovered by the Bremen scientists in Lake Zug in Switzerland. “Our finding opens the possibility that simple unicellular eukaryotes, such as protists, can host energy-providing endosymbionts to complement or even replace the functions of their mitochondria,” says Jon Graf, first author of the study. “This protist has managed to survive without oxygen by teaming up with an endosymbiont capable of nitrate respiration.“ The endosymbiont’s name ‘Candidatus Azoamicus ciliaticola’ reflects this; a ‘nitrogen friend’ that dwells within a ciliate.
An intimate partnership becomes ever closer
So far, it has been assumed that eukaryotes in oxygen-free environments survive through fermentation, since mitochondria require oxygen in order to generate energy. The fermentation process is well documented and has been observed in many anaerobic ciliates. However, microorganisms cannot draw as much energy from fermentation, and they typically do not grow and divide as quickly as their aerobic counterparts.
“Our ciliate has found a solution for this,” says Graf. “It has engulfed a bacterium with the ability to breathe nitrate and integrated it into its cell. We estimate that the assimilation took place at least 200 to 300 million years ago”. Since then, evolution has further deepened this intimate partnership.
The evolution of mitochondria has proceeded in a similar way. „All mitochondria have a common origin,“ explains Jana Milucka. It is believed that more than a billion years ago when an ancestral archaeon engulfed a bacterium, these two started a very important symbiosis: this event marked the origin of the eukaryotic cell. Over time, the bacterium became more and more integrated into the cell, progressively reducing its genome. Properties no longer needed were lost and only the ones that benefitted the host were retained. Eventually, mitochondria evolved, as we know them today. They have their own tiny genome as well as a cell membrane, and exist as so-called organelles in eukaryotes. In the human body, for example, they are present in almost every cell and supply them – and thus us – with energy.
“Our endosymbiont is capable of performing many mitochondrial functions, even though it does not share a common evolutionary origin with mitochondria,” says Milucka. “It is tempting to speculate that the symbiont might follow the same path as mitochondria, and eventually become an organelle”.
A chance encounter
It is actually amazing that this symbiosis has remained unknown for so long. Mitochondria work so well with oxygen – why shouldn't there be an equivalent for nitrate? One possible answer is that no one was aware of this possibility and so no one was looking for it. Studying endosymbioses is challenging, as most symbiotic microorganisms cannot be grown in the laboratory. However, the recent advances in metagenomic analyses have allowed us to gain a better insight into the complex interaction between hosts and symbionts. When analyzing a metagenome, scientists look at all genes in a sample. This approach is often used for environmental samples as the genes in a sample cannot be automatically assigned to the organisms present (see box). This means that scientists usually look for specific gene sequences that are relevant to their research question. Metagenomes often contain millions of different gene sequences and it is quite normal that only a small fraction of them is analyzed in detail
Originally, the Bremen scientists were also looking for something else. The Research Group Greenhouse Gases at the Max-Planck-Institute for Marine Microbiology investigates microorganisms involved in methane metabolism. For this, they have been studying the deep-water layers of Lake Zug. The lake is highly stratified, which means that there is no vertical exchange of water. The deep-water layers of Lake Zug thus have no contact with surface water and are largely isolated. That is why they contain no oxygen but are rich in methane and nitrogen compounds, such as nitrate.
Metagenomic Analysis
The genome is the sum of genes in an organism. In classical genome analysis, the genetic material of a particular species is sequenced using established methods. Every year scientists publish hundreds of different genomes in special data bases. However, the classical approach does not work well for environmental samples. For example, in water samples there is a mixture of many different organisms, and gene sequences cannot be readily assigned to a particular organism. Therefore, metagenomic analyses are used. The term metagenome means the sum of genomes of different species of organisms.
The major challenge using metagenomic analyses is to assign the mixture of genomes to individual species. This problem can be illustrated with an example: Just imagine that the books of several different authors got hopelessly mixed up. The texts are only available in many fragments. The task is to restore the original books. As every author uses a specific way to write it is possible to reconstruct the original texts using statistical analyses of the fragments. This is how the analysis of a metagenome works. A mathematical algorithm helps to solve these puzzles. Fragments (gene sequences) can be put together, can be read and afterwards be reconstructed as individual genomes.
While looking for methane munching bacteria with genes for nitrogen conversion, Graf came across an amazingly small gene sequence that encoded the complete metabolic pathway for nitrate respiration. “We were all stunned by this finding and I started comparing the DNA with similar gene sequences in a database,” says Graf. But the only similar DNA belonged to that of symbionts that live in aphids and other insects. “This didn’t make sense. How would insects get into these deep waters? And why?,” Graf remembers. The scientists of the research group started guessing games and betting.
Not alone in the dark lake

In the end, one thought prevailed: The genome must belong to a yet-unknown endosymbiont. To verify this theory, members of the research team undertook several expeditions to Lake Zug in Switzerland. With the help of the local cooperation partner Eawag they collected samples to look specifically for the organism that contains this unique endosymbiont. In the lab, the scientists fished out various eukaryotes out of the water samples with a pipette. At last, using a gene marker, it was possible to visualize the endosymbiont and identify its protist host.
A final excursion one year ago was supposed to bring final certainty. It was a difficult undertaking in the middle of winter. Stormy weather, dense fog and time pressure due to first news about Coronavirus as well as a possible lockdown made the search in the big lake even more difficult. Nonetheless, the scientists succeeded in retrieving several samples from the deep water and bringing them to Bremen. These samples brought them final confirmation of their theory. “It is nice knowing that they are down there together,” says Jana Milucka. “Normally, these ciliates eat bacteria. But this one let one alive and partnered up with it”.
Many new questions
This finding provokes many exciting new questions. Are there similar symbioses that have existed much longer and where the endosymbiont has already crossed the boundary to an organelle? If such symbiosis exists for nitrate respiration, does it also exist for other compounds? How did this symbiosis, which has existed since 200 to 300 million years, end up in a post-glacial lake in the Alps that only formed 10,000 years ago? Moreover: “Now that we know what we are looking for, we found the endosymbiont’s gene sequences all around the world,” says Milucka. In France, as well as in Taiwan, or in East African lakes that in part are much older than Lake Zug. Does the origin of this symbiosis lie in one of them? Or did it start in the ocean? These are the questions that the research group wants to investigate next.
Report in the Nature Podcast
Jana Milucka gives a detailed report of the discovery of the new symbiosis in the Nature Podcast. Here is the link: https://www.nature.com/nature/podcasts
Original publication
Jon S. Graf, Sina Schorn, Katharina Kitzinger, Soeren Ahmerkamp, Christian Woehle, Bruno Huettel, Carsten J. Schubert, Marcel M. M. Kuypers, Jana Milucka: Anaerobic endosymbiont generates energy for ciliate host by denitrification. Nature, 2021
Participating institutions
- Max Planck Institute for Marine Microbiology, Bremen, Germany
- Max-Planck-Genome-Centre Cologne, Germany
- Eawag, Surface Waters-Research and Management, Kastanienbaum, Switzerland
Please direct your queries to:
Group Leader
Greenhouse Gases Research Group
MPI für Marine Mikrobiologie
Celsiusstr. 1
D-28359 Bremen
Deutschland
|
Room: |
3128 |
|
Phone: |

