Page path:
- Press Office
- Press releases 2012
- 04.05.2012 Algal blooms in the North Sea
04.05.2012 Algal blooms in the North Sea
To most people, algal blooms are an annoyance, which interferes with their summer days by the sea. In the coastal zone of temperate regions a spring algal bloom is not a sign of excessive nutrient input, but most of all a consequence of the more intense solar irradiation in spring (Figure 1). Hence, spring algal blooms in these waters are a natural phenomenon (Figure 2). When algal blooms end the algae die and their remnants constitute an important nutrient supply for the whole ecosystem. This process is essential e.g. for the offshore abundance of fish. But what exactly happens if an algal bloom ends? Hanno Teeling and Bernhard Fuchs and their colleagues from the Max Planck Institute in Bremen provide a surprising and very detailed answer, along with their coauthors from the Alfred Wegener Institute for Marine and Polar Research and the University of Greifswald. They examined an algal bloom in the North Sea and were able to identify the role that the microorganisms play in the degradation of algal remnants and to gain insight into the degradation processes. And they discovered that some of these processes proceed differently than hitherto assumed (Figure 3). Their results were just published in the journal Science.
Figure 1: The algae bloom in the German Bight spreads along the East and West Frisian coast. The satellite image from 2011 also shows the sediment discharge of the rivers Elbe and Weser that mix with the algae bloom. The island Helgoland is encircled in yellow. © NASA images courtesy Jeff Schmaltz, MODIS Rapid Response Team, Goddard Space Flight Center.
For their analyses the scientists filtrated several hundreds of liters of seawater on a regular basis for almost a year off the station ”Kabeltonne”, a long-term station of the Biologische Anstalt Helgoland that is part of the Alfred Wegener Institute. Hanno Teeling from the Max Planck Institute says: “Pelagic microorganisms, the so called bacterioplankton, are critical for the breakdown of the dead algal biomass. Especially the dynamic succession in the bacterioplankton caught our attention. Specialized bacterial populations accompany different phases of the algal bloom”. Processes within the bacterial population control the degradation of the algae, as the scientists could show.
His colleague Bernhard Fuchs who has been investigating the diversity and bacterioplankton composition for many years at the Max Planck Institute, adds: “For the first time we performed a high resolution analysis of the microbial community at genus level. We could not only identify the bacterial groups but also their functional tools, the enzymes, that are involved in the breakdown of the algal bloom”.
His colleague Bernhard Fuchs who has been investigating the diversity and bacterioplankton composition for many years at the Max Planck Institute, adds: “For the first time we performed a high resolution analysis of the microbial community at genus level. We could not only identify the bacterial groups but also their functional tools, the enzymes, that are involved in the breakdown of the algal bloom”.
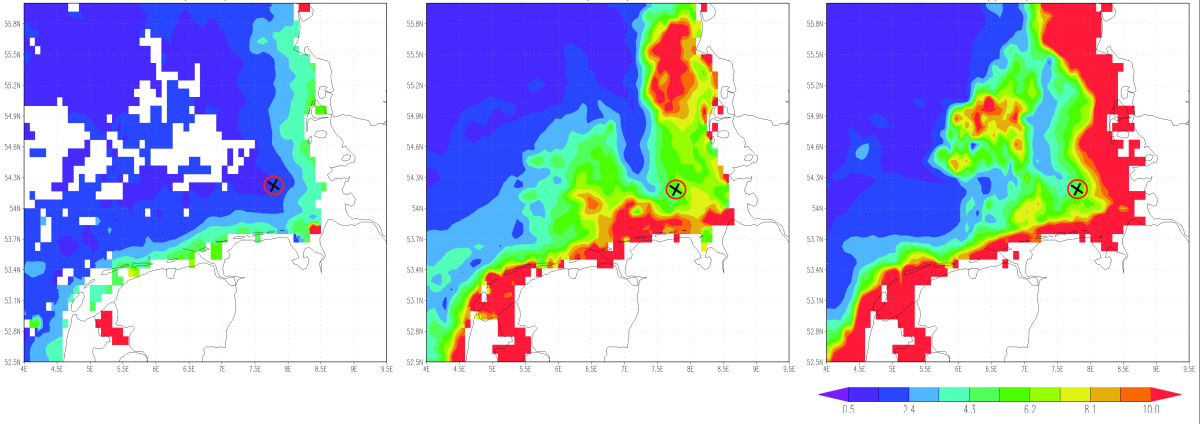
Figure 2: The chlorophyll a content - an indicator for the presence of algae in the water - increased from February (left) to April (right) in the year 2009, in which the samples were taken. At the beginning of April the algae bloom, which consisted mainly of diatoms, reached its maximum and was subsequently degraded. Helgoland is situated in the center of the bloom (black and red mark). © Giovanni online data system, developed and maintained by the NASA GES DISC
The scientists used a novel combination of techniques for their analyses. They determined the identity of the microorganisms by CARD-FISH, an in situ technology that can be applied directly to environmental samples (Figure 4). Additionally, they probed the bacterial population during and after the algal bloom by short sequences of a phylogenetic marker gene (16S rRNA pyrotag analyses). “By using a combination of metagenome and metaproteome analyses we succeeded to detect the active key enzymes in complex environmental samples. This allows us to infer the role of the respective bacterial groups from their metabolic function”, explains Thomas Schweder from the University of Greifswald. “This was only possibly by the computer-controlled integration of all data. For that task we used bioinformatics ”, as Frank Oliver Glöckner from the Max Planck Institute states. In the early phase of the algal bloom the scientists encountered a variety of enzymes for the degradation of complex algal carbohydrates such as laminarin. At a later stage transport proteins for peptides, short protein units, and transporters for the growth limiting nutrient phosphate and simple sugar components dominated the enzymatic cocktail. Noteworthy was the high portion of certain transport proteins, the TonB-dependent transporters that can transport larger molecules directly into the interior of the cells. This discovery may disprove the conventional acceptance that long-chained molecules need to be broken up into smaller components before the cell can take them up. The TonB-transporter may enable the Flavobacteria, one of the dominating bacterial groups, to couple the assimilation and degradation and thus to gain a competitive advance towards other bacterial groups. At the end of the bloom the bacteria increasingly produced sulfatases that cleave sulfate esters from algae carbohydrates hard to decompose and thus allow the complete degradation of these substances. Hence, the scientists discovered a bacterial population in the algae bloom that did not only differ in its composition but also in its function from the bacterial community found in crystal clear, remote open waters.
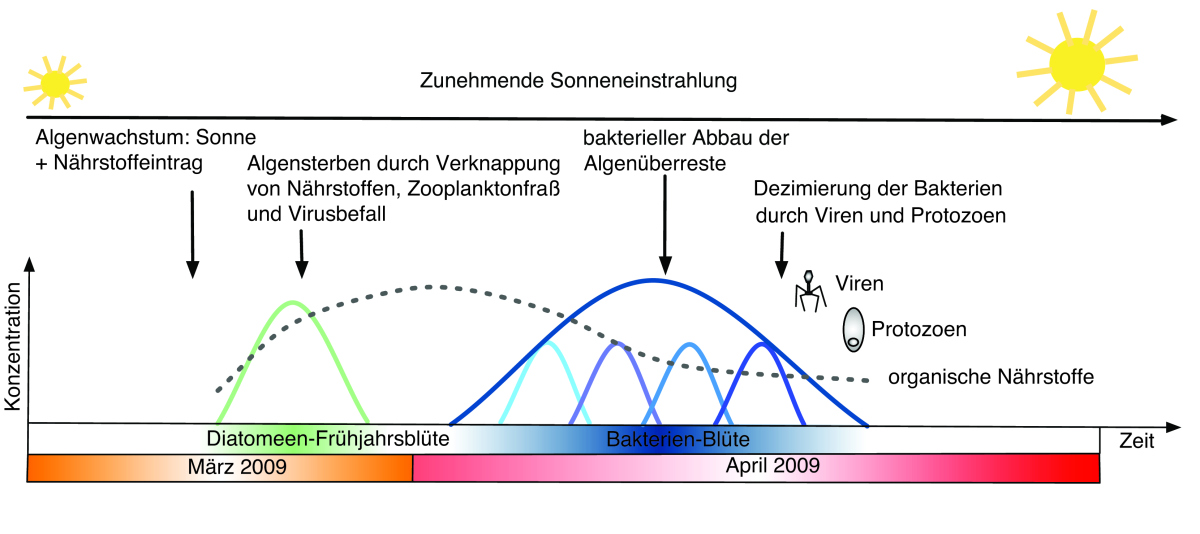
Figure 3: Scheme depicting the algae-bacteria interactions in spring 2009. It takes barely two month for the algae bloom to develop and to be degraded by a subsequent bacterioplankton bloom. The formation of the algae bloom is strongly triggered by the increasing solar radiation in spring. Not only bacteria, but also viruses and zooplankton participate in the degradation of the bloom. It is also the viruses and protozoa, which eventually terminate the bacterial bloom. © H. Teeling/R. Dunker
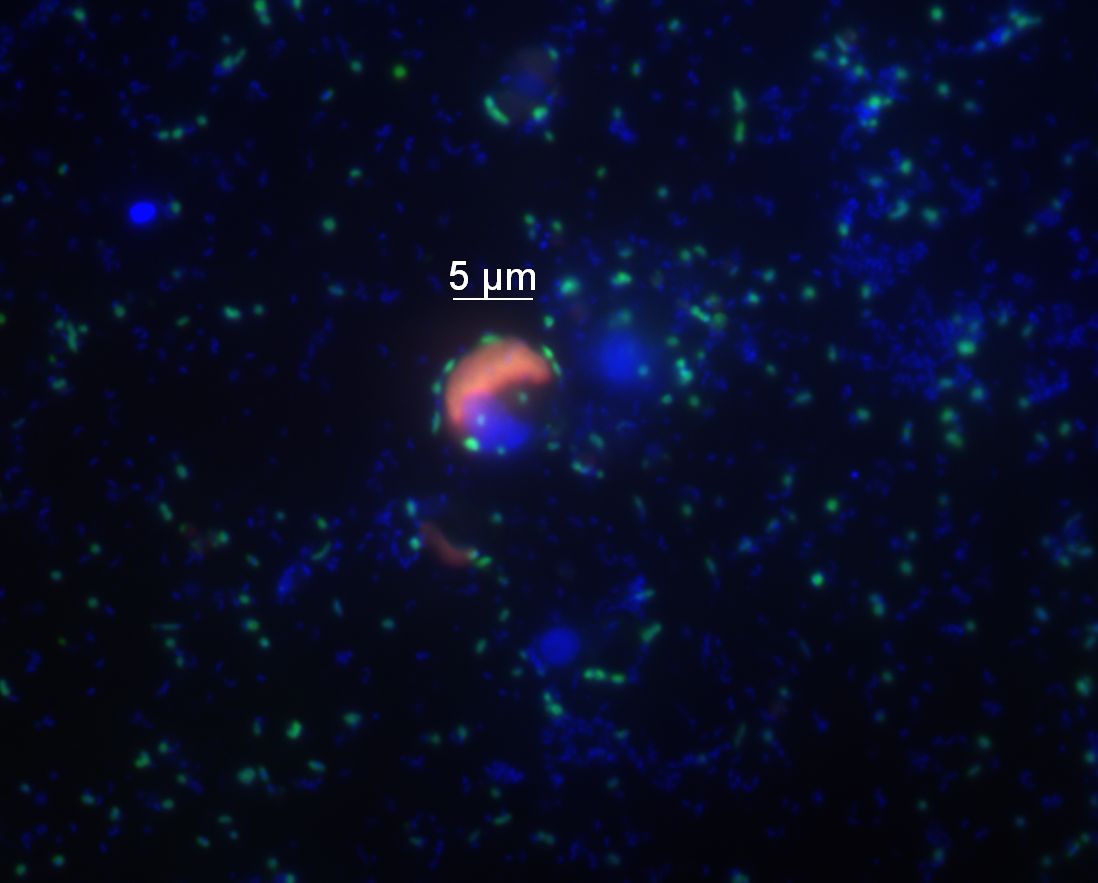
Figure 4: Flavobacteria (green) occur both free living and attached to microalgae in the North Sea. In this image some flavobacterial cells accumulated around a microalgae cell (chloroplasts in red). All other cells are shown in blue. The technique of Fluorescence in situ Hybridization that was used in this image allows for the staining and visualization of bacteria in the intact sample without the need of prior cultivation. © P. Gomez-Perreira/B. Fuchs
The results of the study may help the scientists to resolve the so-called plankton paradox: How can so many plankton species coexist in a seemingly homogeneous habitat without competing for nutrients in a way that eliminates certain species? Rudolf Amann, Director of the Max Planck Institute explains: ”The secret at the level of the microorganisms is the heterogeneity of the microniches that the different groups inhabit. Thus, the specialized populations complement each other in the degradation of the organic matter.”
Rita Dunker
For further information please contact
Dr. Hanno Teeling [Bitte aktivieren Sie Javascript]
Dr. Bernhard Fuchs [Bitte aktivieren Sie Javascript]
Prof. Dr. Rudolf Amann [Bitte aktivieren Sie Javascript]
Prof. Dr. Thomas Schweder [Bitte aktivieren Sie Javascript]
Or the public relation office
Rita Dunker [Bitte aktivieren Sie Javascript]
Manfred Schlösser [Bitte aktivieren Sie Javascript]
Original article
Substrate-controlled succession of marine bacterioplankton populations induced by a phytoplankton blom, 2012. H. Teeling, B. M. Fuchs, D. Becher, C. Klockow, A. Gardebrecht, C. M. Bennke, M. Kassabgy, S. Huang, A. J. Mann, J. Waldmann, M. Weber, A. Klindworth, A. Otto, J. Lange, J. Bernhardt, C. Reinsch, M. Hecker, J. Peplies, F. D. Bockelmann, U. Callies, G. Gerdts, A. Wichels, K. H. Wiltshire, F. O. Glöckner, T. Schweder, and R. Amann. Science, 336: 608-611.
Involved institutions
Max Planck Institute for Marine Microbiology, Bremen
Institute of Marine Biotechnology e.V., Greifswald
Jacobs University Bremen, Bremen
Alfred Wegener Institute for Polar and Marine Research, Biologische Anstalt
Helgoland, Helgoland
Institute for Microbiology, Ernst-Moritz-Arndt University, Greifswald
Pharmaceutical Biotechnology, Ernst-Moritz-Arndt University, Greifswald
DECODON GmbH, Greifswald
Ribocon GmbH, 28359 Bremen
Helmholtz-Zentrum Geesthacht, Center for Materials and Coastal Research, Geesthacht
Rita Dunker
For further information please contact
Dr. Hanno Teeling [Bitte aktivieren Sie Javascript]
Dr. Bernhard Fuchs [Bitte aktivieren Sie Javascript]
Prof. Dr. Rudolf Amann [Bitte aktivieren Sie Javascript]
Prof. Dr. Thomas Schweder [Bitte aktivieren Sie Javascript]
Or the public relation office
Rita Dunker [Bitte aktivieren Sie Javascript]
Manfred Schlösser [Bitte aktivieren Sie Javascript]
Original article
Substrate-controlled succession of marine bacterioplankton populations induced by a phytoplankton blom, 2012. H. Teeling, B. M. Fuchs, D. Becher, C. Klockow, A. Gardebrecht, C. M. Bennke, M. Kassabgy, S. Huang, A. J. Mann, J. Waldmann, M. Weber, A. Klindworth, A. Otto, J. Lange, J. Bernhardt, C. Reinsch, M. Hecker, J. Peplies, F. D. Bockelmann, U. Callies, G. Gerdts, A. Wichels, K. H. Wiltshire, F. O. Glöckner, T. Schweder, and R. Amann. Science, 336: 608-611.
Involved institutions
Max Planck Institute for Marine Microbiology, Bremen
Institute of Marine Biotechnology e.V., Greifswald
Jacobs University Bremen, Bremen
Alfred Wegener Institute for Polar and Marine Research, Biologische Anstalt
Helgoland, Helgoland
Institute for Microbiology, Ernst-Moritz-Arndt University, Greifswald
Pharmaceutical Biotechnology, Ernst-Moritz-Arndt University, Greifswald
DECODON GmbH, Greifswald
Ribocon GmbH, 28359 Bremen
Helmholtz-Zentrum Geesthacht, Center for Materials and Coastal Research, Geesthacht