- Research & Instruments
- How we study - Our instruments and methods
How we study - Our instruments and methods
≈ 60 weeks
is how long Tramper has already been autonomously on its way. Set down on the ocean floor at a depth of several thousand metres, this robot moves slowly forward and regularly measures the oxygen content in the seafloor. Its data provides information on local element cycles: Microorganisms break down organic material from the remains of dead plants and animals. The more organic material sinks down from the upper water layers, the more oxygen is consumed.
> 1 year
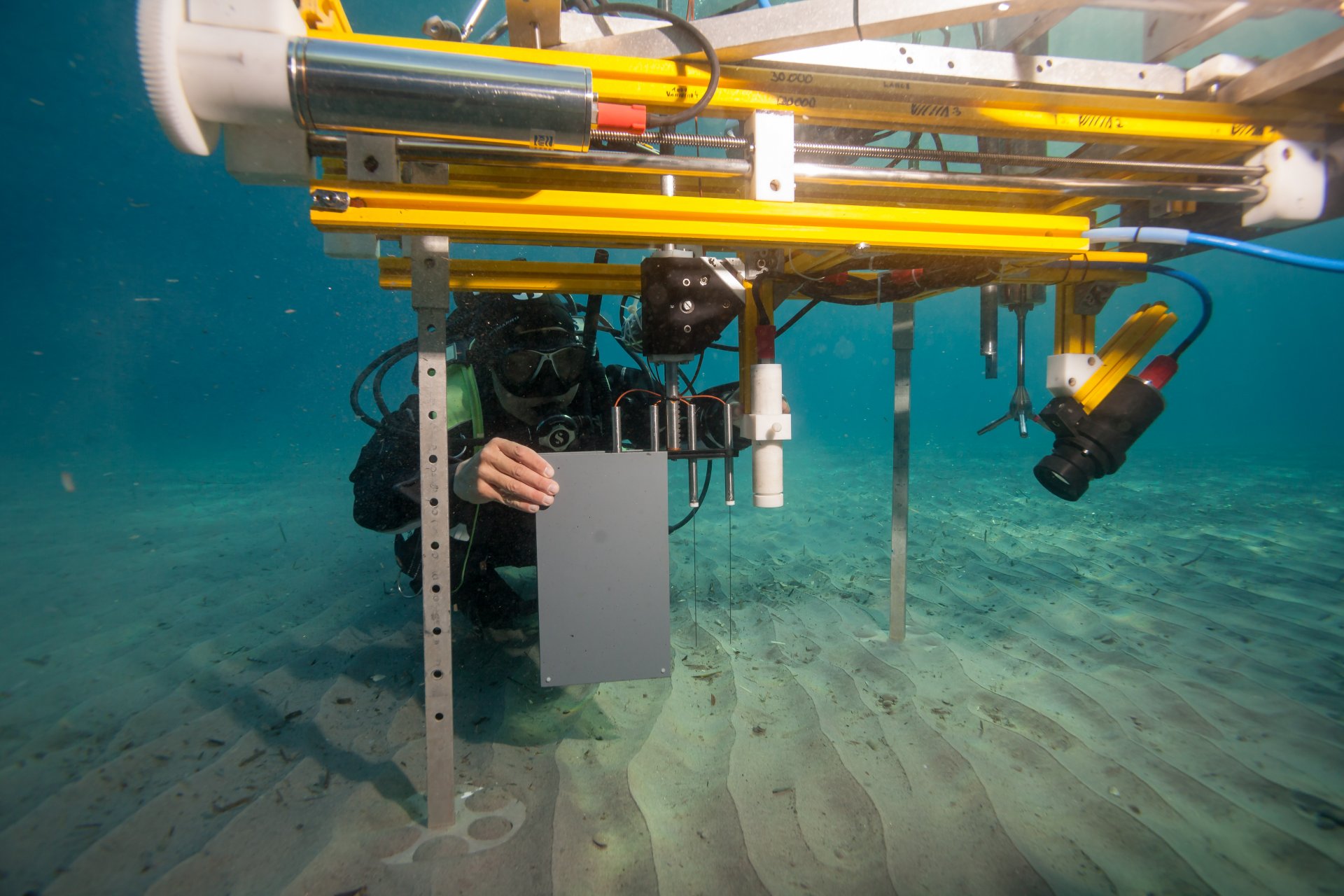
Good things take time. This is also often the case for research equip-ment. For example, it took a lot of employees and more than a year before Lance-A-Lot was ready for deployment. And we continue to redesign and develop it further.
20 to 50 micrometres
Razor-sharp measurements: Our needle-like microsensors are generally finer than a human hair. Using these extremely thin glass capillaries, we can study the conditions in the smallest spaces and in the immediate environment of a single bacterium. For the sensors to be perfectly suited to our scientific needs, we build them ourselves.
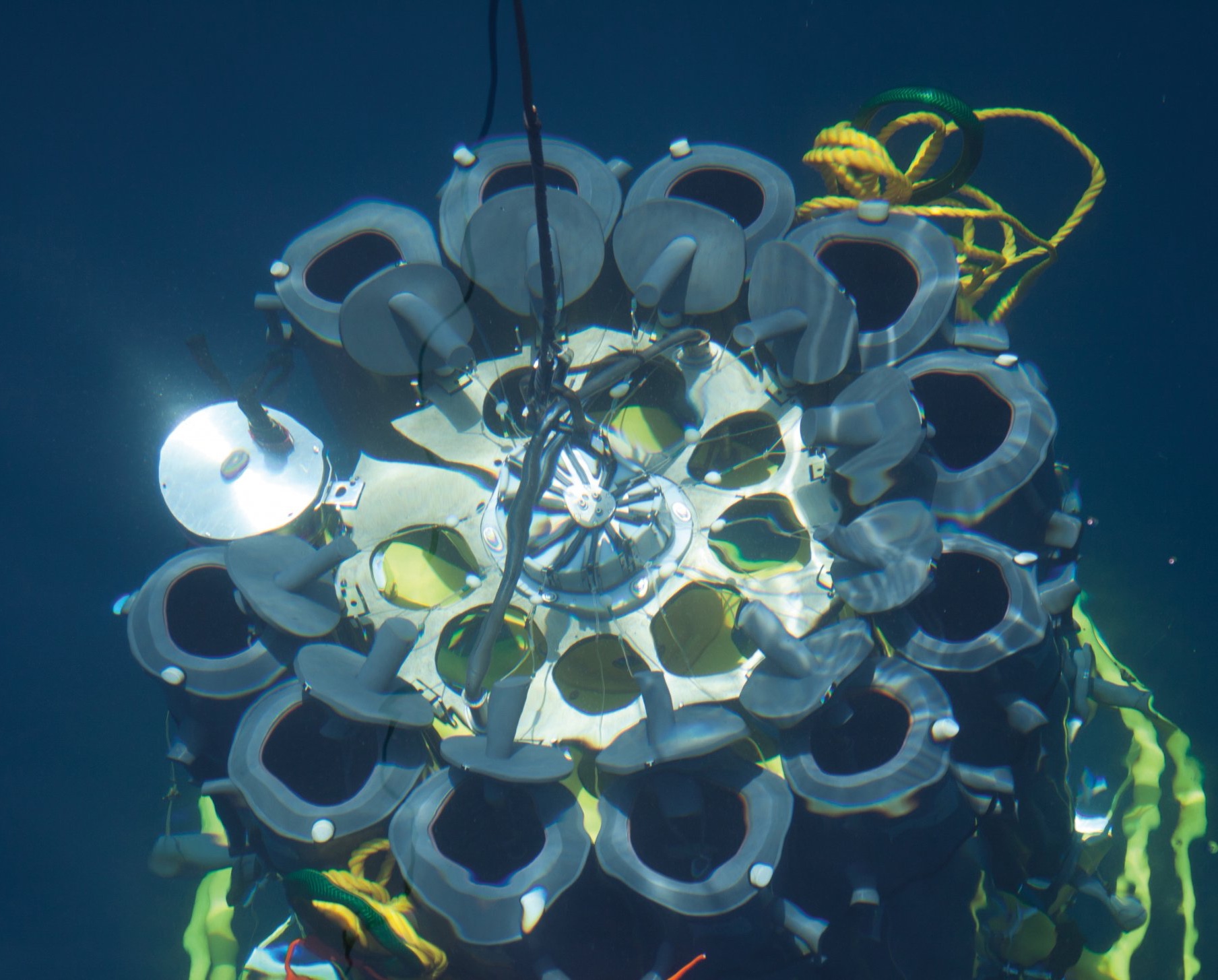
Thousands of kilograms
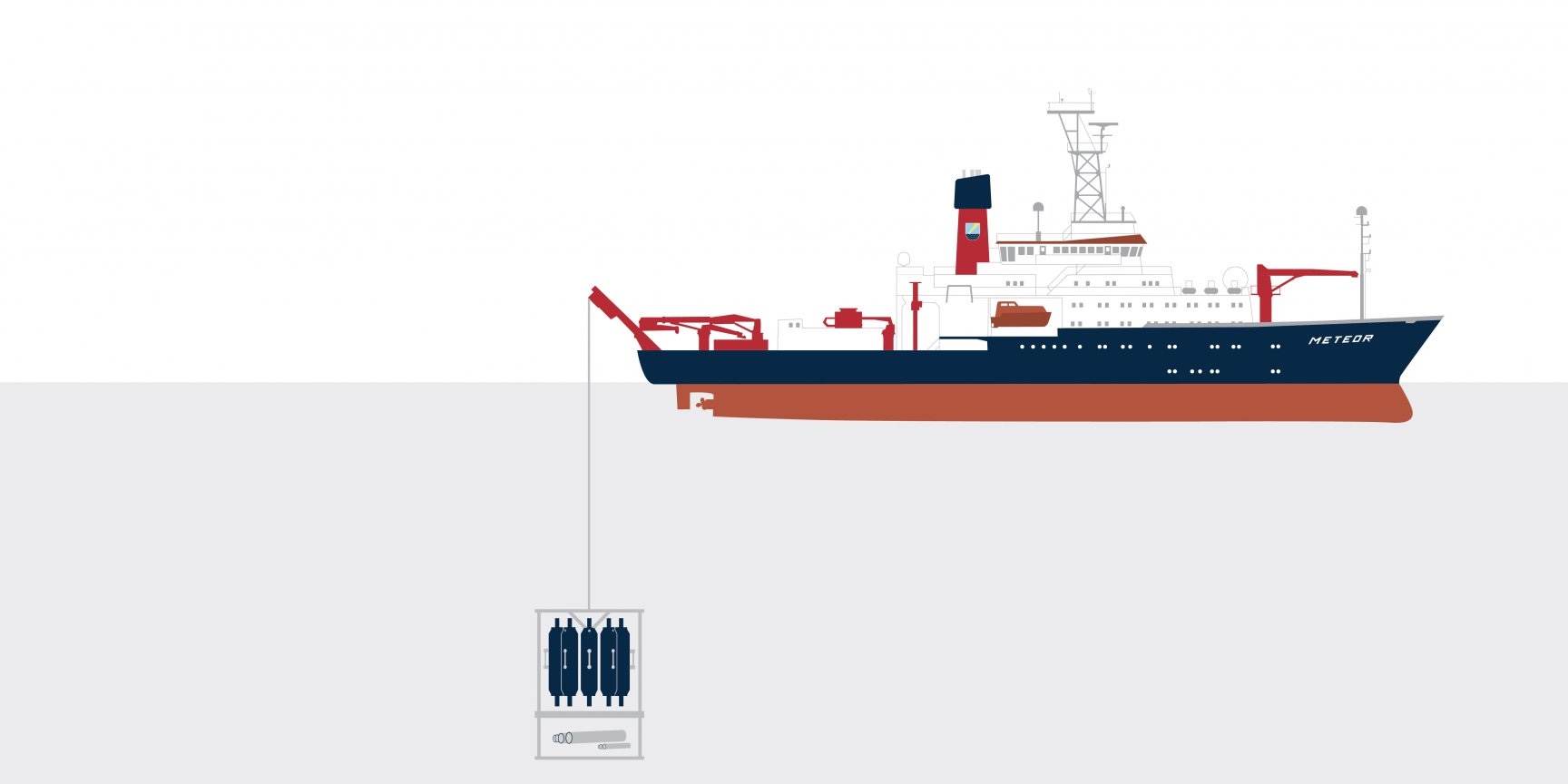
How does research equipment get safely from the ship into the water and down to the seafloor? With a strong cable, for example. Which is itself a heavyweight: A 1.8 centimetre thick cable weighs 1.2 kilograms per metre. That adds up: To reach the bottom of a deep-sea trench, we must deploy almost 10,000 kilos of cable!
Our Research Facilities
At the Max Planck Institute for Marine Microbiology, we use many different types of equipment - from simple buckets to common light microscopes and highly complex high-tech machines. A unique feature of our institute is the large variety of different imaging techniques. We have a Nano SIMS, a MALDI-Imaging mass spectrometer, various electron microscopes and many different techniques to analyse genes and genoms. All under one roof.
Researching deeper thanks to technology: The world’s oceans end when they hit continents, but these narrow coastal strips, the direct point of contact between the sea and the land, are just one of many microbial habitats we study. To reach the microcosmos in both the open and the deep ocean, we need our largest piece of equipment: Ships take us to our research locations. Even samples and measured data can only be acquired using sophisticated equipment. And once we have collected our water and sediment samples, it is important to make the microorganisms within them visible and, even more difficult, the internal structures within the microorganisms. Our challenge, then, is not just the research itself, but also how it can best be accomplished. Designing, building and developing new equipment and technologies is therefore an inseparable part of our scientific work.
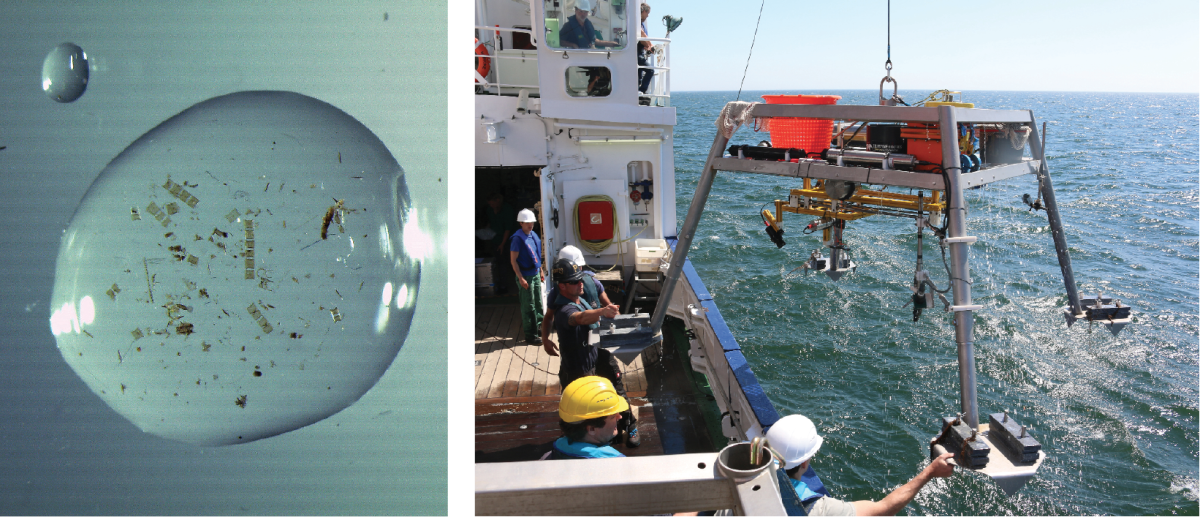
Lance-A-Lot: Triple investigation
Just like the legendary knight of King Arthur’s Round Table, Lance-A-Lot has special powers: It simultaneously measures the flow velocity, the topography of the seafloor and oxygen levels in the sand. Where there are ripples, for example, oxygen and nutrients can penetrate deeper into the sediment than on a smooth surface. The microorganisms under ripples are therefore better supplied with nutrients. The flow rate in turn affects what and how much the bacteria breathe in the sand and what remains in the water.
Super Zoom: Insights beyond resolution
The majority of microbes are not visible to the naked eye. The microscope helps us to see individual cells. But we want to look even closer. Using high-resolution microscopes, we can even recognize the structures inside a microorganism. With the correct technology, even individual genes can be made visible! These objects are at the limits of resolution – the distance that two things must at least be apart to still be separated optically. Technical tricks make this possible: For example, we can make fluorescent molecules within the cells flash. Subsequent computer analyses generate an image that shows us exactly where the individual molecules are within a sample. This allows us to image cell structures approximately five to ten times more accurately than with conventional fluorescence microscopy alone.
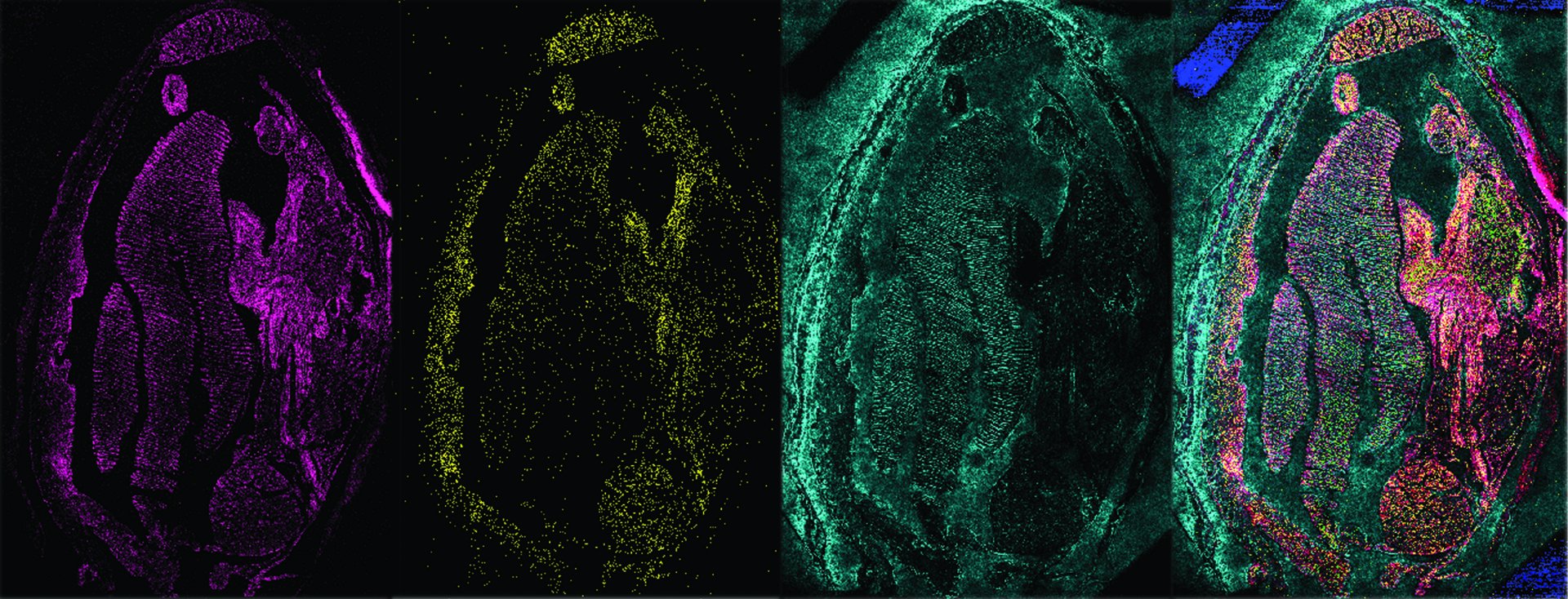
FISH: Identifying individuals
Microorganisms can barely be distinguished from each other based on their appearance. Nevertheless, every cell has its own fingerprint that is typical for the species – its genetic material. A case for FISH, Fluorescence In Situ Hybridisation. It selectively visualizes certain sections of the genetic material of individual cells. Under the microscope, these cells are then made to glow. FISH images of microorganisms look like a starry sky, but one with many different colours. Our question is: What is shining where? With FISH, we can precisely identify the organisms in our samples.
NanoSIMS: Viewed in detail
NanoSIMS stands for Nanoscale Secondary Ion Mass Spectrometer. This mass spectrometer possesses a special optical system that allows enormous spatial resolution: We can observe things as small as 50 nanometres – that is, a twentieth of a millionth of a metre. For example, we investigate the structures and processes in a single cell and can virtually watch it working. This is quite unique: There are only around 20 of these instruments worldwide. Ours was the first to be used to address ecological problems.
Bioinformatics: An ocean of knowledge
The genetic material of microbes, their DNA, conceals a lot of information about their lifestyle and abilities. To analyze the DNA, we would have to culture and investigate these tiny creatures in the laboratory – but this is not possible for most microorganisms. We therefore analyze environmental samples with all the microbes they contain. An enormous hodgepodge! We keep a clear view thanks to bioinformatics: For example, we analyze short pieces of DNA strands and then compile them to long sequences on the computer. Even the complete genomes of individual species can be reconstructed in this way. We gain in-depth knowledge about individual microbes from bioinformatics, but also of entire ecosystems.

HyperDiver: Mapping reefs
Coral reefs are beautiful – and extremely sensitive. To protect them, we need to know them well and measure their health. But how? With our HyperDiver! It uses the colours of the corals: Their colour spectra are different depending on which species they belong to and their condition. HyperDiver monitors a wide range of wavelengths – many more than the human eye can perceive – and thus registers the condition of the reef. Image recognition and self-learning software evaluate the data. A diver can record up to 40 square metres of reef per minute with centimetre accuracy. A spin-off team from our institute has developed a user-friendly version of this technology and is currently commercializing it (here is the link to the start-up from the Max Planck Institute, PlanBlue).
Mass spectrometry: Tracking individual molecules
The coexistence of microbes with each other or animals is all about molecules. From nutrition to communication, proteins, lipids and the like are almost always involved. We make these molecules visible and gain insights into the processes within cells and between cells. For example, our MALDI-MSI can visualize the distribution of thousands of molecular compounds, without having to stain or label them beforehand. This molecular microscope uses the molecular mass of the individual compounds. We then use the data to generate images from which we can identify which compound is where and how abundant it is. Other mass spectrometers use isotopes, which are different forms of the same element. For example, ratios of carbon isotopes reveal how microorganisms take up carbon dioxide – and thus provide insights into the global carbon cycle.