- Press Office
- Press releases 2021
- The earthworm in new light
The earthworm in new light
Earthworms experience constant chemical interactions with bacteria, fungi, plants and small invertebrates across soil ecosystems. Even within their tissues, earthworms harbor symbiotic microbes and small animal parasites that trigger internal metabolic responses such as innate immunity. To reveal the fundamental processes that enable animal–microbe symbioses to form and persist, we have to study their metabolic interactions in situ. By combining novel imaging techniques, a team of researchers around Benedikt Geier from the Max Planck Institute for Marine Microbiology (MPIMM) in Bremen, Germany, has now succeeded in imaging the exciting variety of interactions that take place inside the earthworm. This allows us to observe them in a completely new light.
The chemistry of the worm in 3D
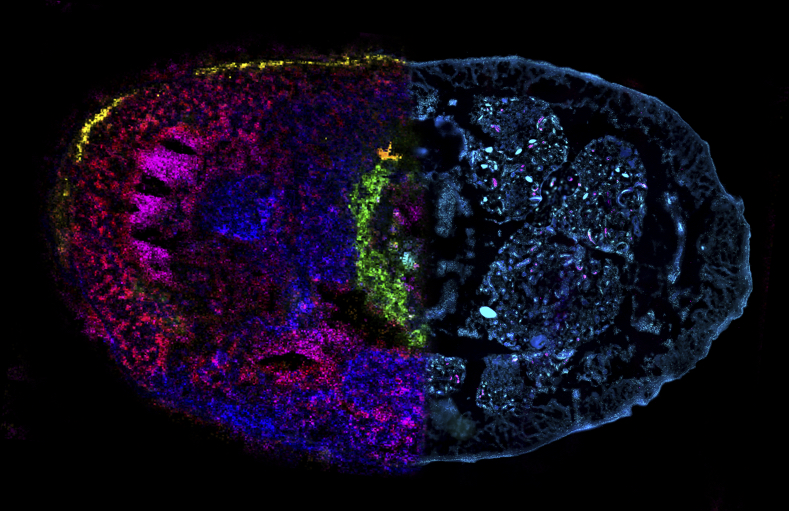
Our understanding of the chemical interactions between small animals and the microorganisms that live inside their bodies is extremely limited. This gap of knowledge has its origin in a methodical challenge: To study the metabolic interactions in symbiosis we have to know who is producing which metabolic product where in the body. The core of this problem, however, is not only to image how molecules are distributed at the micrometer scale. Also, the chemical images are almost impossible to interpret without knowing if a tissue sample is healthy, diseased or infected with beneficial or pathogenic microbes or even animal parasites.
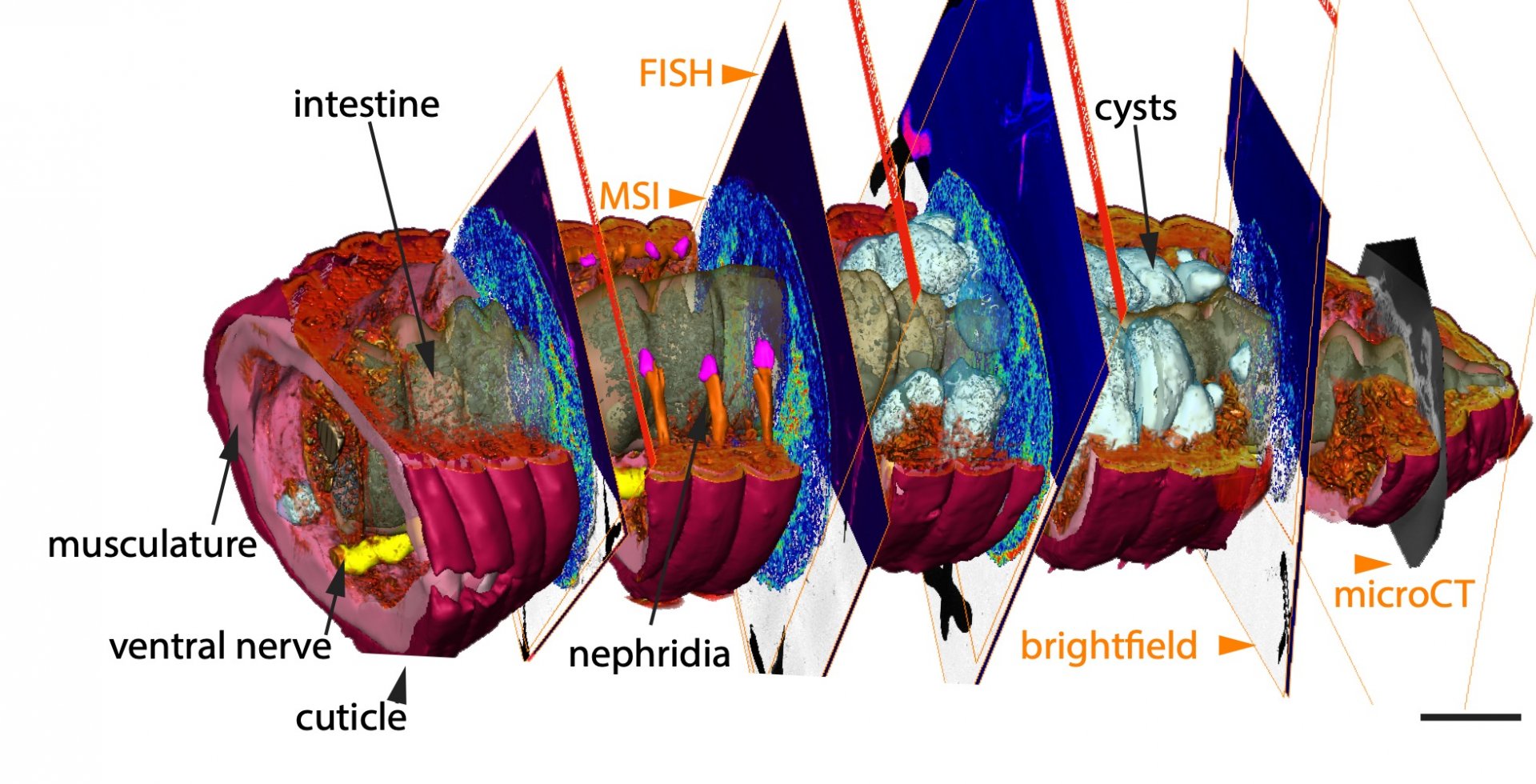
The new combination of high-resolution imaging techniques can offer a solution to this problem, as the research team now shows. “In our study, we introduce chemo-histo-tomography, a special three-dimensional imaging approach of the chemistry and anatomy of millimeter-sized animals and their parasites at a cellular level,” says Benedikt Geier, first author of the paper. “This method offers a new strategy to visualize the most fundamental processes – metabolic interactions – in small animal symbioses. It allows us to spatially assign metabolic producs to the animal host and its microbial partners at the micrometer scale.“
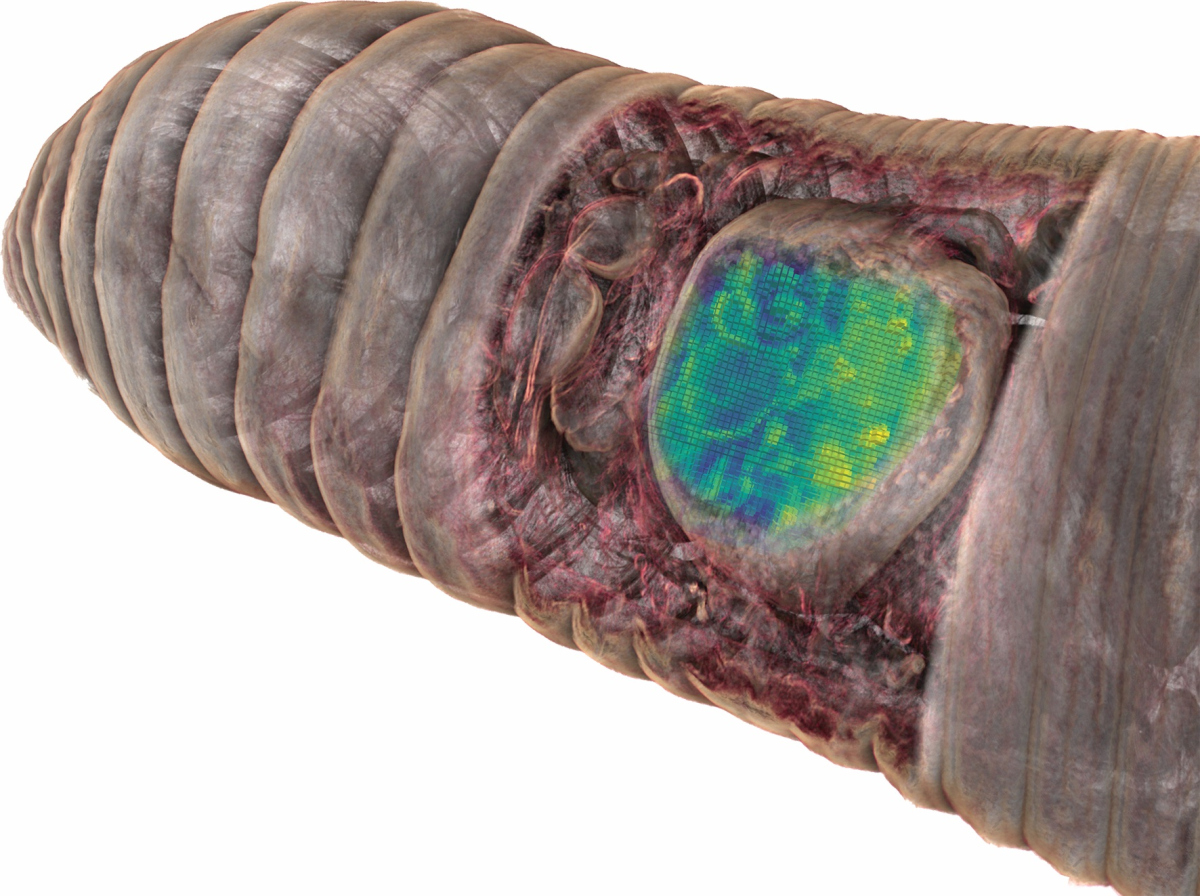
Chemo-histo-tomography (CHEMHIST) combines chemical imaging of metabolites based on MALDI mass spectrometry imaging with the microanatomy of the same animal that is recorded with micro-computed X-ray tomography. The latter is a non-invasive approach allowing X-ray imaging of the 3D histology, and can reach subcellular resolution. For metabolite imaging, MALDI mass spectrometry techniques have become a state-of-the-art tool to visualize micrometer-scale, natural distributions of metabolites. This enables a spatial assignment between chemical profiles and their production site and possibly to their producer.
“This advance allowed us to take an earthworm from the environment and create a 3D atlas of its chemical and physical interactions with the microorganisms naturally occurring inside its tissues,” says Manuel Liebeke, leader of the Research Group Metabolic Interactions at the MPIMM and senior author of the study. “However, we were interested in more than just the biology of the earthworm. Our objective was to also make CHEMHIST applicable to animals directly sampled from their natural habitat, which can be essential for symbiosis research.” The resolution of CHEMHIST outperforms existing similar methods developed for medical research in mice by up to two orders of magnitude. This may also lead into new avenues for research on insects or corals, which are key models for symbiosis research, both on land and in water.
The whole is more than the sum of the parts
Using the combination of different in situ imaging techniques through CHEMHIST, the researchers at the MPIMM in Bremen uncovered metabolites – products of the metabolism – in the earthworm that could shed light on how it chemically defends itself against parasites and how these, in turn, protect themselves against the earthworm's immune response. However, to even recognize that the chemical image data were metabolites of parasites in earthworm tissue, the 3D anatomical model was indispensable. A high-resolution micro-computer tomograph at the German Electron Synchrotron in Hamburg first made it possible to identify the parasites in the tissue.
Notably, metabolic interactions between animals and their microbes are not restricted to symbiotic tissues. Along the gut-brain axis, microbial metabolites produced in the gut can affect tissues across the host, even reaching the brain. Therefore, extending correlative chemical imaging into 3D approaches can be crucial for capturing the distribution of metabolites involved in symbiotic interactions and thus show how chemical signals from microbes possibly affect crucial processes in their host. Also, the method can be used in many ways: The research team around Liebeke and Geier is currently applying it to deep-sea mussels.
“Since I have specialized in the 3D visualization of invertebrate anatomy for years, it was particularly interesting for me to see the molecules hidden behind the morphological structures,” says Bernhard Ruthensteiner, leader of the section Invertebrate Varia at the Bavarian State Collection of Zoology. The results of the study were made possible by an interdisciplinary collaboration that brought together scientists from the fields of microbiology, zoology, chemistry and physics. It quickly became clear that visualization such as the correlative 3D atlas of the worm facilitate science communication of the data.
Original publication
Benedikt Geier; Janina Oetjen; Bernhard Ruthensteiner; Maxim Polikarpov; Harald Gruber-Vodicka; Manuel Liebeke: Connecting structure and function from organisms to molecules in small animal symbioses through chemo-histo-tomography, PNAS, Juni 2021
Participating institutions
- Max Planck Institute for Marine Microbiology, Bremen, Germany
- Bavarian State Collection of Zoology, Munich, Germany
- European Molecular Biology Laboratory, Hamburg Unit c/o German Electron Synchrotron in Hamburg, Germany
- MALDI Imaging Lab, University of Bremen, Germany
Please direct your queries to:
Group leader
MPI for Marine Microbiology
Celsiusstr. 1
D-28359 Bremen
Germany
|
Room: |
3244 |
|
Phone: |