Taxonomy
Director
Managing Director
Department of Molecular Ecology
MPI for Marine Microbiology
Celsiusstr. 1
D-28359 Bremen
Germany
|
Room: |
2221 |
|
Phone: |

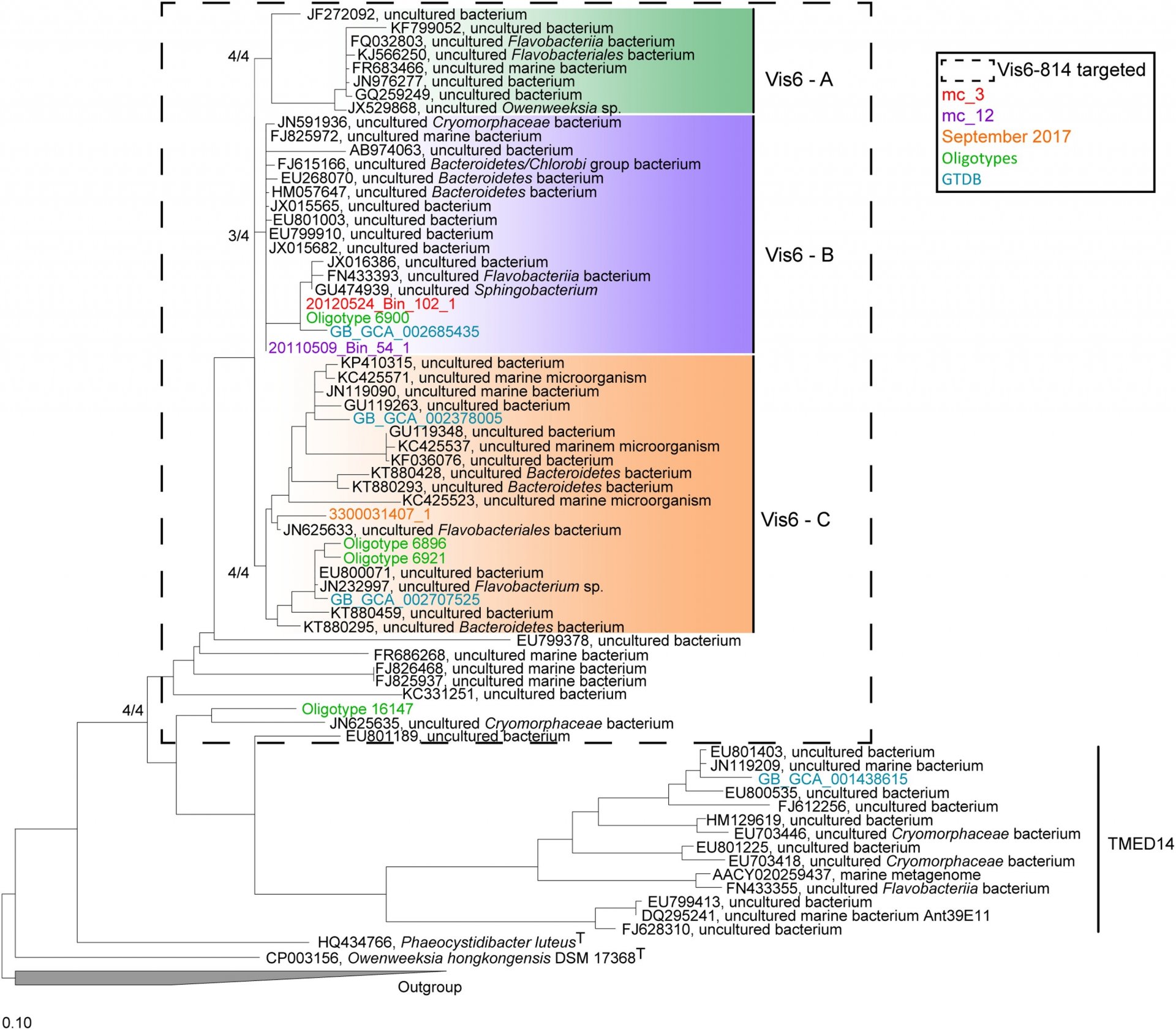
Uncultivated microbes in need of their own taxonomy
Taxonomy encompasses the identification, classification and nomenclature of organisms. As such, taxonomy is a prerequisite for ecology. Only based on accurate taxonomic concepts and methods, can the diversity and composition of complex microbial communities be accurately described and monitored. Our department has a long tradition of developing and applying new taxonomic methods. We have pioneered the in situ identification, quantification and localization of not-yet cultivated bacteria and archaea by fluorescence in situ hybridization (FISH) with rRNA-targeted oligonucleotide probes (Amann et al. 1995; Amann & Fuchs 2008). We have a long-standing collaboration with Wolfgang Ludwig and Ralf Westram, the main developers of the ARB program that is widely used for the reconstruction of 16S rRNA-based phylogenetic trees and for the design of oligonucleotide probes. Emerging from that program, we also have long been the home of the much-used, curated 16S rRNA database SILVA (Quast et al. 2013; Yilmaz et al. 2014). This has now been transferred to the Leibniz Institute DSMZ-German Collection of Microorganisms and Cell Cultures.
Since the majority of microorganisms have not yet been cultivated, there was a need for thresholds to assess whether their 16S rRNA fingerprints indicate the existence of new species, genera, families, orders, classes or even new phyla. We contributed to widely used standards of 16S rRNA-based classification (Yarza et al. 2014). Today, the genomic revolution is enabling very detailed descriptions of new taxa without cultivation, and it is evident that order must be brought to the rather uncontrolled alphanumerical naming of "novel" (Konstantinidis et al. 2017). Single cell genomics and/or metagenome-assembled genomes, often combined with single cell identification by FISH, discloses the true diversity of microorganisms, which is breathtaking, yet certainly not endless (Amann & Rossello-Mora 2016). Currently, others and we have identified a pressing need to do proper taxonomy of abundant and functionally important clades of environmental bacteria and archaea. As such, we have recently described several new marine taxa including the species Candidatus Prosiliicoccus vernus (Francis et al. 2019) and the genus Candidatus Abditibacter (Grieb et al. 2020). However, since the rank of Candidatus is preliminary and the nomenclature lacks priority, others and we are convinced that genomic information needs to be accepted as type material for the permanent description of novel microorganisms (Konstantinidis et al. 2020). Ecologists and taxonomists require a roadmap for naming uncultivaed Archaea and Bacteria (Murray et al. 2020) that is compatible with the current Code of Nomenclature requiring live pure cultures as type material. We believe that this will not hinder the enrichment and cultivation work of others and us that is more needed than ever. We now not only know that something is out there but also can predict what it is doing, characterising its ecology while we wait for cultivation efforts to provide a more detailed inspection. These "ghosts" have names and we know their genomes.